Plot ESM data on unstructured grids with psyplot#
This notebook introduces you to the mapplot function of the package psyplot and its plugin psy_maps.
It is suitable to plot maps from data on unstructured grids like the ones from ICON and FESOM.
We therefore search for the corresponding data in the CMIP6 data pool with intake-esm.
Afterwards, we open a file with xarray and configure the opened xarray dataset as well as psyplot for a map plot.
This Jupyter notebook is meant to run in the Jupyterhub server of the German Climate Computing Center DKRZ. The DKRZ hosts the CMIP data pool including 4 petabytes of CMIP6 data. Please, choose the Python 3 unstable kernel on the Kernel tab above, it contains all the common geoscience packages. See more information on how to run Jupyter notebooks at DKRZ here.
Running this Jupyter notebook in your premise, which is also known as client-side computing, will require that you install the necessary packages
intake, xarray, maplotlib, psyplot, psy_maps
and either download the data or use the opendap_url column of the intake catalog if available.
Learning Objectives#
How to access data on an unstructured grid from the DKRZ CMIP data pool with
intake-esmHow to subset data with
xarrayHow to visualize the results with
matplotlib,psyplotandpsy_maps.
import psyplot.project as psy
import matplotlib as mpl
import xarray as xr
import intake
We open a swift catalog from dkrz cloud which is accessible without authentication.
# Path to master catalog on the DKRZ server
#dkrz_catalog=intake.open_catalog(["https://dkrz.de/s/intake"])
#
#only for the web page we need to take the original link:
parent_col=intake.open_catalog(["https://gitlab.dkrz.de/data-infrastructure-services/intake-esm/-/raw/master/esm-collections/cloud-access/dkrz_catalog.yaml"])
list(parent_col)
['dkrz_cmip5_archive',
'dkrz_cmip5_disk',
'dkrz_cmip6_cloud',
'dkrz_cmip6_disk',
'dkrz_cordex_disk',
'dkrz_dyamond-winter_disk',
'dkrz_era5_disk',
'dkrz_monsoon_disk',
'dkrz_mpige_disk',
'dkrz_nextgems_disk',
'dkrz_palmod2_disk']
col=parent_col["dkrz_cmip6_disk"]
col.df["uri"]=col.df["uri"].str.replace("lustre/","lustre02/")
In this example, we aim at plotting the Sea Surface Temperature tos of the upper boundary of the liquid ocean, including temperatures below sea-ice and floating ice shelves from the earth system model AWI-CM-1-1-MR.
We therefore search for tos in the catalog for monthly frequency. We only use one realization r1i1p1f1 of one experiment only.
tos=col.search(source_id="AWI-CM-1-1-MR",
experiment_id="ssp370",
variable_id="tos",
table_id="Omon",
member_id="r1i1p1f1")
tos.df["uri"].to_list()[0]
'/work/ik1017/CMIP6/data/CMIP6/ScenarioMIP/AWI/AWI-CM-1-1-MR/ssp370/r1i1p1f1/Omon/tos/gn/v20181218/tos_Omon_AWI-CM-1-1-MR_ssp370_r1i1p1f1_gn_201501-202012.nc'
We now open the file on the mistral lustre file system. Note that if you work remotely, you could try to use opendap_url instead of path.
dset = xr.open_dataset(tos.df["uri"].to_list()[0],
decode_cf=True,
chunks={"time":1},
lock=False)
dset
<xarray.Dataset>
Dimensions: (time: 72, bnds: 2, ncells: 830305, vertices: 16)
Coordinates:
* time (time) datetime64[ns] 2015-01-16T12:00:00 ... 2020-12-16T12:00:00
lat (ncells) float64 dask.array<chunksize=(830305,), meta=np.ndarray>
lon (ncells) float64 dask.array<chunksize=(830305,), meta=np.ndarray>
Dimensions without coordinates: bnds, ncells, vertices
Data variables:
time_bnds (time, bnds) datetime64[ns] dask.array<chunksize=(1, 2), meta=np.ndarray>
tos (time, ncells) float32 dask.array<chunksize=(1, 830305), meta=np.ndarray>
lat_bnds (ncells, vertices) float64 dask.array<chunksize=(830305, 16), meta=np.ndarray>
lon_bnds (ncells, vertices) float64 dask.array<chunksize=(830305, 16), meta=np.ndarray>
Attributes: (12/39)
frequency: mon
Conventions: CF-1.7 CMIP-6.2
creation_date: 2018-12-18T12:00:00Z
data_specs_version: 01.00.27
experiment: ssp370
experiment_id: ssp370
... ...
parent_experiment_id: historical
parent_mip_era: CMIP6
parent_source_id: AWI-CM-1-1-MR
parent_time_units: days since 1850-1-1
parent_variant_label: r1i1p1f1
activity_id: ScenarioMIP AerChemMIPIn order to make tos plottable, we set the following configuration.
The
CDI_grid_typeis a keyword forpsyplot. It must match the grid type of the source model.Coordinates are not fully recognized by
xarrayso that we have to add some manually (version from Dec 2020).
dset["tos"]["CDI_grid_type"]="unstructured"
coordlist=["vertices_latitude", "vertices_longitude", "lat_bnds", "lon_bnds"]
dset=dset.set_coords([coord for coord in dset.data_vars if coord in coordlist])
The following is based on the psyplot example. We set a resoltion for the land sea mask lsm and a color map via cmap.
psy.rcParams['plotter.maps.xgrid'] = False
psy.rcParams['plotter.maps.ygrid'] = False
mpl.rcParams['figure.figsize'] = [10, 8.]
def plot_unstructured():
iconplot11=psy.plot.mapplot(
dset, name="tos", cmap='rainbow',
clabel=dset["tos"].description,
stock_img=True, lsm='50m')
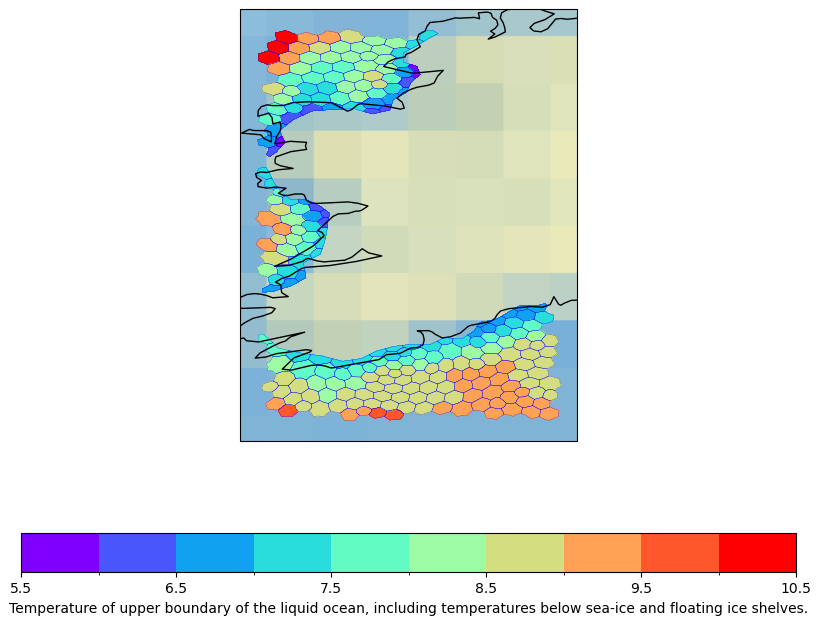
We now do the same with a smaller subset to highlight the fine resolution and the structure of the AWI ocean model FESOM.
We first subset the data because otherwise plotting takes too long. We choose indices of dimensions with the xarray function isel. We select a slice of two time steps and focus on a region Ireland. We have to save the data to an intermediate file test.nc because otherwise we receive an error.
dset["lat"]=dset.lat.load()
dset["lon"]=dset.lon.load()
dset2 = dset.isel(time=slice(1,2)).where( (dset.lon > -10. ) &
(dset.lon < 50. ) &
(dset.lat > 40. ) &
(dset.lat < 70. ), drop=True).drop("time_bnds")
dset2
<xarray.Dataset>
Dimensions: (time: 1, ncells: 28844, vertices: 16)
Coordinates:
* time (time) datetime64[ns] 2015-02-15
lat (ncells) float64 69.74 69.16 69.17 69.72 ... 69.8 67.23 42.17
lat_bnds (ncells, vertices) float64 dask.array<chunksize=(28844, 16), meta=np.ndarray>
lon (ncells) float64 -8.645 -9.653 -5.883 -6.825 ... 47.55 47.76 18.49
lon_bnds (ncells, vertices) float64 dask.array<chunksize=(28844, 16), meta=np.ndarray>
Dimensions without coordinates: ncells, vertices
Data variables:
tos (time, ncells) float32 dask.array<chunksize=(1, 28844), meta=np.ndarray>
Attributes: (12/39)
frequency: mon
Conventions: CF-1.7 CMIP-6.2
creation_date: 2018-12-18T12:00:00Z
data_specs_version: 01.00.27
experiment: ssp370
experiment_id: ssp370
... ...
parent_experiment_id: historical
parent_mip_era: CMIP6
parent_source_id: AWI-CM-1-1-MR
parent_time_units: days since 1850-1-1
parent_variant_label: r1i1p1f1
activity_id: ScenarioMIP AerChemMIPdset2.to_netcdf("test.nc")
dset2.close()
dset=xr.open_dataset("test.nc",
decode_cf=True)
dset["tos"]["CDI_grid_type"]="unstructured"
coordlist=["vertices_latitude", "vertices_longitude", "lat_bnds", "lon_bnds"]
dset=dset.set_coords([coord for coord in dset.data_vars if coord in coordlist])
psy.plot.mapplot(
dset, name="tos", cmap='rainbow',
lonlatbox='Ireland',
clabel=dset["tos"].description,
stock_img=True,
lsm='50m',
datagrid=dict(c='b', lw=0.2)).show()
/envs/lib/python3.11/site-packages/cartopy/io/__init__.py:241: DownloadWarning: Downloading: https://naturalearth.s3.amazonaws.com/50m_physical/ne_50m_coastline.zip
warnings.warn(f'Downloading: {url}', DownloadWarning)
dset.close()
Used data#
We acknowledge the CMIP community for providing the climate model data, retained and globally distributed in the framework of the ESGF. The CMIP data of this study were replicated and made available for this study by the DKRZ.”
